By Yu Jianbin, People’s Daily
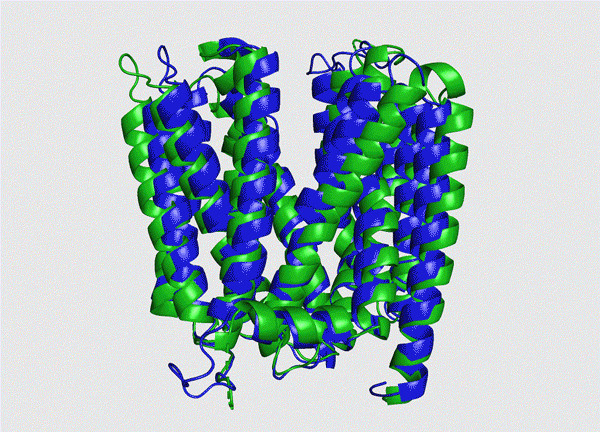
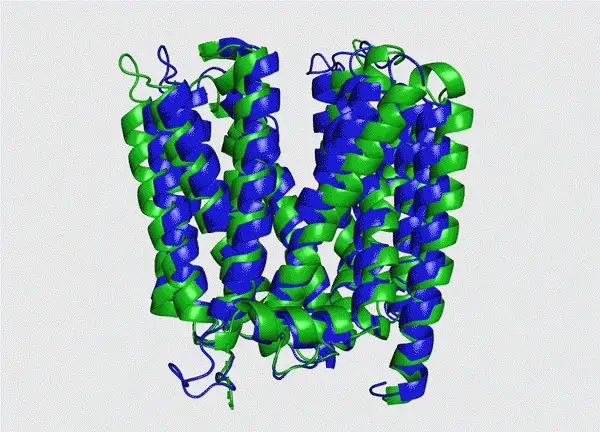
Photo shows the structure of a type of protein. The green part is the real structure, while the blue part is the prediction. (Photo from the website of Tianrang)
Shanghai-based Chinese artificial intelligence (AI) startup Tianrang recently announced that its independently developed deep-learning protein folding prediction platform had delivered remarkable performance in the 14th round of the Critical Assessment of Structure Prediction, ranking among the best of its same kind in the world. It took only 16 seconds for the platform to predict the protein chains of 400 amino acids.
Proteins are major functional molecules that play a variety of roles in cells. For instance, as a catalyzing enzyme, they adjust metabolism and carry metabolic substances to form cytoskeletons. Besides, they also join the immunologic, cell differentiation and apoptosis process.
To decode the functions of proteins, a basic element composing life forms, is fundamental to uncovering the secrets of biological phenomena.
Xue Guirong, founder of Tianrang, introduced that proteins must fold into particular structures in order to perform their functions. Only a few types of proteins are functional when they are not folding. The functions of proteins are determined by their three-dimensional structures. Once the structures are damaged, the proteins lose their functions.
Many diseases are caused by abnormal structures of important proteins. Therefore, the studies on protein structures lead to better understanding of proteins’ functions, thus stimulate improvement in healthcare, food sustainability and innovative biotechnology, and promote the development of life science, R&D of drugs and synthetic biology.
The observation and analysis of protein structure have always been a fascinating topic in life science which many scientists are studying. However, the study is difficult and costly, and facing limited progress.
There are three major traditional methods to observe protein structures - magnetic resonance imaging, radiology and cryogenic electron microscopy. These methods need huge trial and error and expensive devices, and normally take years to complete the study of a single structure. In addition, these experimental methods are not capable enough to reveal some important protein structures, and more bioinformation technologies and computational biology approaches are needed.
However, usual computer software could not take the astonishingly massive computing volume of protein structure, and even supercomputers are not able to do that. Therefore, protein structure prediction has become an important branch of structural biology, and researchers predict spatial structure of proteins based on amino acid sequence through relevant AI algorithm they have developed.
“AI is showing surprising decision-making capabilities in dealing with complicated systematic problems, from defeating the world champion of the Go game, to managing urban traffic. The prediction of protein structures, though a biological issue, is also a problem in complicated scenarios where AI can demonstrate its huge potential in handling basic science research. That’s something we don’t want to miss,” said Xue.
Such a project is very precious, as it has made innovations in inter-discipline studies, industry, basic science, AI algorithms and engineering capability, Xue noted.
The recent progress indicated that the application of AI in the studies of protein structure can decode some structures that are not able to be analyzed through traditional observation methods, and the results are reliable and very close to the fact. Such AI-powered structure prediction is expected to become a sharp tool of scientists and accelerate the development of research in life science.
At present, the prediction of single protein folding is just a start. Proteins often function as compounds in pairs or groups, and the structures of many protein compounds are still a mystery.
Xue believes that the adaptability and precision of AI algorithms shall be further improved, so as to make contributions to revealing how proteins interact with each other and help mankind find new and accurate ways to cure diseases.
Proteins are major functional molecules that play a variety of roles in cells. For instance, as a catalyzing enzyme, they adjust metabolism and carry metabolic substances to form cytoskeletons. Besides, they also join the immunologic, cell differentiation and apoptosis process.
To decode the functions of proteins, a basic element composing life forms, is fundamental to uncovering the secrets of biological phenomena.
Xue Guirong, founder of Tianrang, introduced that proteins must fold into particular structures in order to perform their functions. Only a few types of proteins are functional when they are not folding. The functions of proteins are determined by their three-dimensional structures. Once the structures are damaged, the proteins lose their functions.
Many diseases are caused by abnormal structures of important proteins. Therefore, the studies on protein structures lead to better understanding of proteins’ functions, thus stimulate improvement in healthcare, food sustainability and innovative biotechnology, and promote the development of life science, R&D of drugs and synthetic biology.
The observation and analysis of protein structure have always been a fascinating topic in life science which many scientists are studying. However, the study is difficult and costly, and facing limited progress.
There are three major traditional methods to observe protein structures - magnetic resonance imaging, radiology and cryogenic electron microscopy. These methods need huge trial and error and expensive devices, and normally take years to complete the study of a single structure. In addition, these experimental methods are not capable enough to reveal some important protein structures, and more bioinformation technologies and computational biology approaches are needed.
However, usual computer software could not take the astonishingly massive computing volume of protein structure, and even supercomputers are not able to do that. Therefore, protein structure prediction has become an important branch of structural biology, and researchers predict spatial structure of proteins based on amino acid sequence through relevant AI algorithm they have developed.
“AI is showing surprising decision-making capabilities in dealing with complicated systematic problems, from defeating the world champion of the Go game, to managing urban traffic. The prediction of protein structures, though a biological issue, is also a problem in complicated scenarios where AI can demonstrate its huge potential in handling basic science research. That’s something we don’t want to miss,” said Xue.
Such a project is very precious, as it has made innovations in inter-discipline studies, industry, basic science, AI algorithms and engineering capability, Xue noted.
The recent progress indicated that the application of AI in the studies of protein structure can decode some structures that are not able to be analyzed through traditional observation methods, and the results are reliable and very close to the fact. Such AI-powered structure prediction is expected to become a sharp tool of scientists and accelerate the development of research in life science.
At present, the prediction of single protein folding is just a start. Proteins often function as compounds in pairs or groups, and the structures of many protein compounds are still a mystery.
Xue believes that the adaptability and precision of AI algorithms shall be further improved, so as to make contributions to revealing how proteins interact with each other and help mankind find new and accurate ways to cure diseases.
 Menu
Menu
 Progress made in AI-powered prediction of protein structures
Progress made in AI-powered prediction of protein structures